A Novel Macrophage Containing Organotypic Small Intestinal Tissue for Modeling Gut Inflammation
- TR Number: 969
- Keywords: EpiIntestinal, SMI-200-FT, Macrophages, TLR4, lipopolysaccharide; LPS, NOD1, C12-iE-DAP, NOD2, L18-MDP, gene analysis, chemokines, chemokine receptors, FC receptors, co-stimulatory molecules, interferons, HLA, IL-6, IL-8, TNF-a, caspase 5, CCL20, , CCL3, CCL3L3, CCL3L1, CEACAM7, CFB complement factor B, CXCL1, CXCL3, CXCL8, IL23A, lipopolysaccharide binding protein
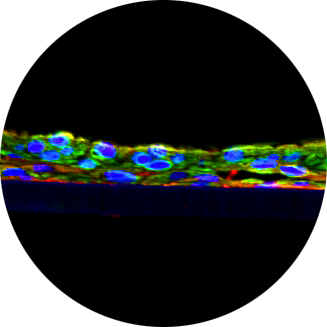
As gatekeepers of intestinal immunehomeostasis, macrophages play a critical role in inflammation in the gut. In this study, were constructed a new macrophage-containing primary cell-based full-thickness small intestinal(SMI+M) tissue model and characterized it for:1) macrophage incorporation (immunohistochemistry, IHC), 2) barrier properties(TEER), and 3)functionality by measuring inflammatory responses following exposure to ligands for TLR4 (lipopolysaccharide;LPS),NOD1(C12-iE-DAP)andNOD2(L18-MDP) either individually or synergistically. For identification of inflammatory responses, we utilized AffymetrixGeneChiparrays. Structurally, the SMI+M tissues have 3D polarity and their morphology and physiological barrier properties mimic those of native in vivo tissues. IHC of SMI+M tissues showed CD14 (macrophagemarker) cells in the underlying fibroblast/collagen layer. Using gene upregulation of >1.9 fold as a cut-off following ligand stimulation, tissues without macrophages (SMI-M) showed fewer upregulated genes (1400genes) compared to SMI+M (4400genes). Furthermore, when gene upregulation levels by ligand-induced SMI+M were compared to stimulated SMI-M, even higher differences in upregulated genes (>5200genes) were noted. In SMI+M, upregulated genes include chemokines, chemokine receptors, FC receptors, co-stimulatory molecules, interferons, and HLA’s which are characteristic of immune cells. When we looked at 15 upregulated genes (>6-fold) in SMI+M, the synergistic effect of ligands in inducing inflammatory gene upregulation was much more pronounced compared to stimulated SMI-M tissues. Furthermore, increased cytokine release of IL-6, IL-8, and TNF-α into the culture medium from ligand exposed SMI+M was also confirmed by BioPlex ELISA. In summary, our results demonstrate that the 3D SMI+M tissue model can serve as an in vitro tool to study the complex cellular interactions manifested during inflammation in the gut microenvironment.